3DbioNet - discussing 3D cell biology
Editors
B. Singh
David A. Turner
Raphaël Lévy
Sandrine Willaime-Morawek
3D Bioprinted In Vitro Metastatic Models via Reconstruction of Tumor Microenvironments (2019)
Fanben Meng, Carolyn M. Meyer, Daeha Joung, Daniel A. Vallera, Michael C. McAlpine, Angela Panoskaltsis-Mortari
DOI: 10.1002/adma.201806899 PubMed: 30663123
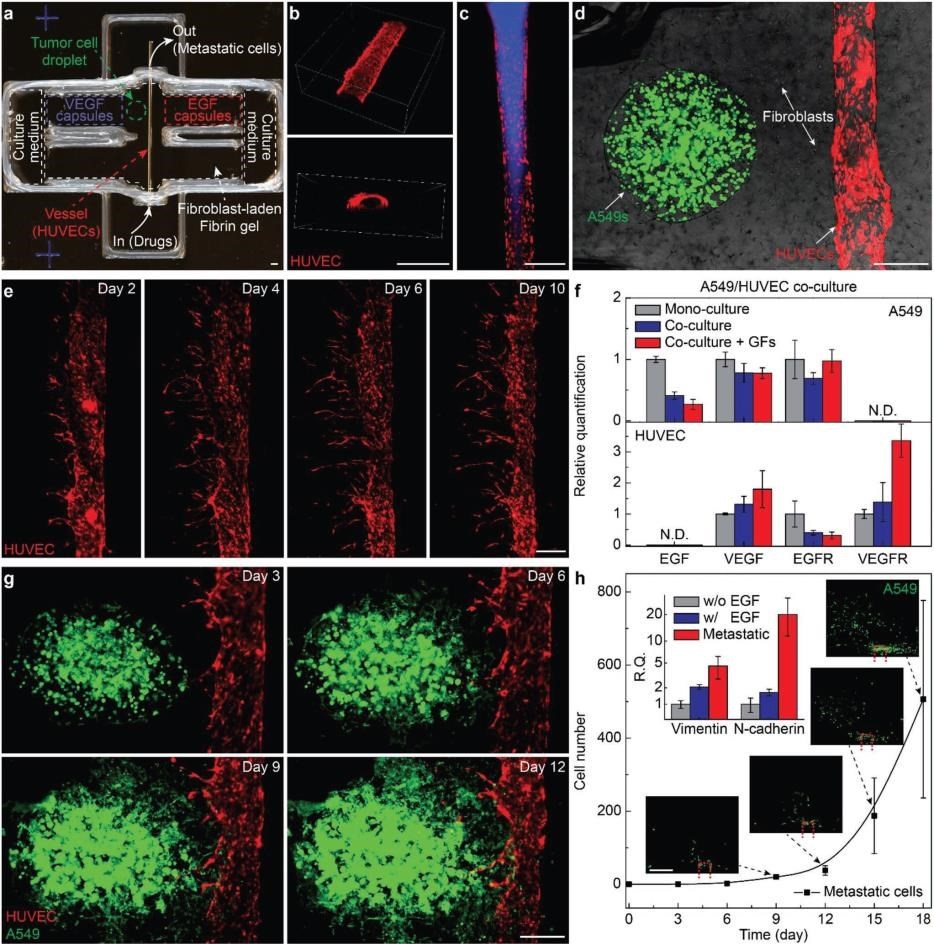
Review by Dr. Priyanka Gupta, University of Surrey, UK. This publication by Meng et al., 2019 attempts to create an in vitro metastasis model of tumour microenvironment (TME) using lung cancer as a model system. The paper employs multiple techniques including 3D bioprinting, stimuli responsive growth factor capsules to create their model. As seen in Figure 1, the TME model designed by the authors contains a 3D printed vascular channel endothelialised with HUVECS to mimic vascularization, 3D bioprinted tumour spheroids in a fibrin gel matrix containing fibroblasts (to mimic tumour stroma) and strategically placed stimuli responsive capsules containing specific growth factors required for tumour cell migration (EGF) and capillary formation by endothelial cells (VEGF). The authors have based their observations and conclusions on imaging and image quantification since it is well established that the currently available methods for biological quantification in 2D is not well suited for complex 3D systems. Additionally, in order to validate the developed model, they have reported the feasibility of using the model to carry out anti-cancer drug screening wherein the in vitro model was able to mimic in vivo observation to a large extent.
Figure 1: 3D printed in vitro tumor models mimicking metastatic dissemination. Schematic image of the integration of tumor cells, endothelial cell-lined vascular conduits, and biochemical signals within a fibroblastladen fibrin gel to reconstruct tumor microenvironments. Tumor cell invasion of the surrounding matrix and intravasation into the vasculature are mediated by EGF and VEGF gradients, which are dynamically generated by 3D printed programmable release capsules (EGF: epidermal growth factor; VEGF: vascular endothelial growth factor; EGFR: EGF receptor; and VEGFR: VEGF receptor). In our opinion, the following are the positive aspects of the research article:
- Modular approach to the problem: The authors went on to develop each aspect of their model one at a time followed by their characterisation. This kind of a modular approach is important as a process and allows others to pick and choose aspects/modules from the model as per requirement for future research in this area.
- Versatility of the model: The developed model chose components which are generic and are applicable to most cancer types (growth factors, stromal cells and stromal matrix) and makes it possible to use it for other cancer.
- Use of emerging technologies and methods: The researchers have used various emerging engineering technologies like 3D bioprinting, stimuli responsive growth factor capsules, gradient guided cellular migration, targeted therapeutic methods etc which is helpful for other researchers in getting an idea of the available techniques which can be employed in the field of tissue engineering (TE) in general.
- Image analysis based quantification technique: There is a general consensus amongst researchers in the field of TE that most of the biological quantification techniques/ methods are not the best options for quantification in a 3D model. The authors of this paper employs image analysis based quantification of their observations which highlights the feasibility of using image quantification for informed conclusions in 3D systems. The article does raise a couple of questions and problems:
- Choice of growth factors and markers: Although VEGF is well known for its effect of endothelial cells, EGF as an exclusive growth factor for tumour cell migration does raise some questions. As is evident from Figure 3f in the paper, lung cancer cells monoculture shows that the cell line A549 secreted EGF on its own. Thus, there are some questions if the cellular migration observed in the model is due to the externally added EGF or is due to the EGF secreted by the cells themselves. Additionally, using only Vimentin and N-cadherin as markers for metastasis is considered to be an incomplete observation (Figure 3h).
- Conclusion related to the importance of stromal cells (fibroblasts): The authors have concluded in the article that stromal cells do not play any direct role in cellular migration for both cancer and endothelial cells. However, as observed in Figure 3d, the number of fibroblasts present within their model is much less than established in vivo observations wherein very high fibrosis is observed. Hence, this raises questions regarding their observations and conclusion related to the importance of tumour stroma and stromal cells.
- Lack of well written experimental method: The experimental methods and materials sections lacked a detailed note of the methods employed, making it difficult to duplicate this TME model. This is also applicable to the image analysis based quantification methods. A more thorough and well explained methods section would have been appreciated by the research community.
Figure 3: a) Photo of a 3D printed culture chamber for tests of guided tumor cell dissemination. b) Confocal images of the top view (upper panel) and cross section (lower panel) of a representative microchannel lined by HUVECs within a fibrin gel, showing the lumen of the vessel. c) Fluorescence images showing a vessel perfused by fluorescent dye. d) Composite image showing a representative tumor model before laser-triggered rupture of EGF and VEGF capsules. e) Panoramic fluorescence images showing sprouts generated from a main vessel and their extension toward a single direction over time, indicating guided sprouting angiogenesis by VEGF capsules. f) Bar chart showing the expression of EGF, VEGF, EGFR, and VEGFR of A549s (upper panel) and HUVECs (lower panel) when mono cell-cultured (gray) and co-cell-cultured without (blue) and with (red) EGF and VEGF within fibrin gels (normalized by the levels of each mono cell-cultured samples, mean± s.d. n = 3 per group, g) Fluorescence images of a metastatic model on days 3, 6, 9, and 12, showing that A549s approach and enter the vasculature through the fibroblast-laden fibrin gel. h) Plots of the population of disseminated A549s detected in the collection chamber versus time. (mean±s.d., n = 3 per group). Scale bar: 500 μm
Overall in conclusion, this article brings to the forefront emerging techniques useful for 3D tissue engineering, especially in the area of 3D in vitro models and adds valuable information to the field of in vitro TME models. There are some gaps in the current paper which requires future studies in this area.
Subjects
A peeriodical is a lightweight virtual journal with you as the Editor-in-chief, giving you complete freedom in setting editorial policy to select the most interesting and useful manuscripts for your readers. The manuscripts you will evaluate and select are existing publications—preprints and papers. Thus, a peeriodical replicates all the functions of a traditional journal, including discovery, selection and certification, except publication itself.
Why set up a peeriodical? The traditional journal has changed remarkably little in centuries and many people feel that scientific publishing is stuck in a rut, subject to a corporatist drift, and is not serving science optimally. The advent of preprints in many fields beyond those served by the ArXiv is liberating the dissemination of research, but most other journal functions have not been replaced effectively. Now you—all researchers—have the opportunity to select and certify research according to your own criteria. We expect peeriodical subject matters and editorial policies to be extremely varied. Some peeriodicals may wish to target narrow domains, while others will adopt a generalist approach. Some peeriodicals will be inclusive, focusing on discovery, whereas others may aim to enforce stringent quality criteria, prioritising certification. The point is that all approaches are permitted and supported—we hope you will innovate! You can create multiple peeriodicals. It will be users and readers who decide which peeriodicals they find useful and interesting. Users can sign up to receive alerts from any peeriodical they wish.
A peeriodical has one or more editors. Anybody can set-up a peeriodical and either operate it alone or invite colleagues to form an editorial board or community. The editors can select "manuscripts"—existing papers or preprints—to consider, either spontaneously or through suggestions from other researchers, including of course the authors. Note that there is no obligation that the manuscript be recent; for instance, we expect that some peeriodicals could focus on underappreciated classics. After all, predictions about scientific impact are generally more accurate for the past than the future. If the editors wish, they can solicit reviews for the manuscript via the Peeriodicals interface. Reviews will be published and the referees will have the option of posting anonymously or signing their review. Editors may decide at any time to accept, reject or comment on the manuscript, taking into account the comments received. They may of course suggest improvements to the manuscript or underlying study. If they justify their decision, their editorial decision will also be published.
How will Peeriodicals fit into the publishing landscape? We see them as a space without entry barriers in which researchers can innovate and explore new approaches to scientific dissemination, in parallel to the traditional publishing industry. There are related and complementary initiatives, notably the overlay journals promoted by Tim Gowers, exemplified by Discrete Analysis, but also Science Open Collections, PLoS Channels, the APPRAISE initiative and Peer Community in... Each of these projects has their own specificities and goals. Nobody yet knows exactly what the future will look like, but we strongly believe that we are about to experience a period of rapid evolution in the dissemination of science and we hope that Peeriodicals will inspire and help you to share your imagination and expertise with the whole research community.
For those starting a peeriodical, you will discover that the hardest part is building up an audience. Unfortunately, we can't yet guarantee you the exposure you would get from a paper in a glamour journal. Reviews with scientific content will be mirrored on PubPeer, offering an audience through the PubPeer browser and Zotero extensions. However, it will be largely up to you to run your publicity, most likely through social media. We are on Twitter (@PEERIODICALS) and will of course help out as we can.
Get started now by requesting an invitation with the link in the top right menu.